We cannot ignore the possibility of another pandemic, especially with the potential for more infectious and fatal disease outbreaks. Pandemic prevention is essential to protect the health and well-being of the American people. The article below from the Boston Globe mentions Dr. Mark C. Poznansky and Dr. Michael V. Callahan of the Infectious diseases department at Massachusetts General Hospital, who both urge for a definite need from national institutions to prepare an organized response for the next pandemic.
Customizing T Cell-Based Immunotherapies in a ‘SNAP’
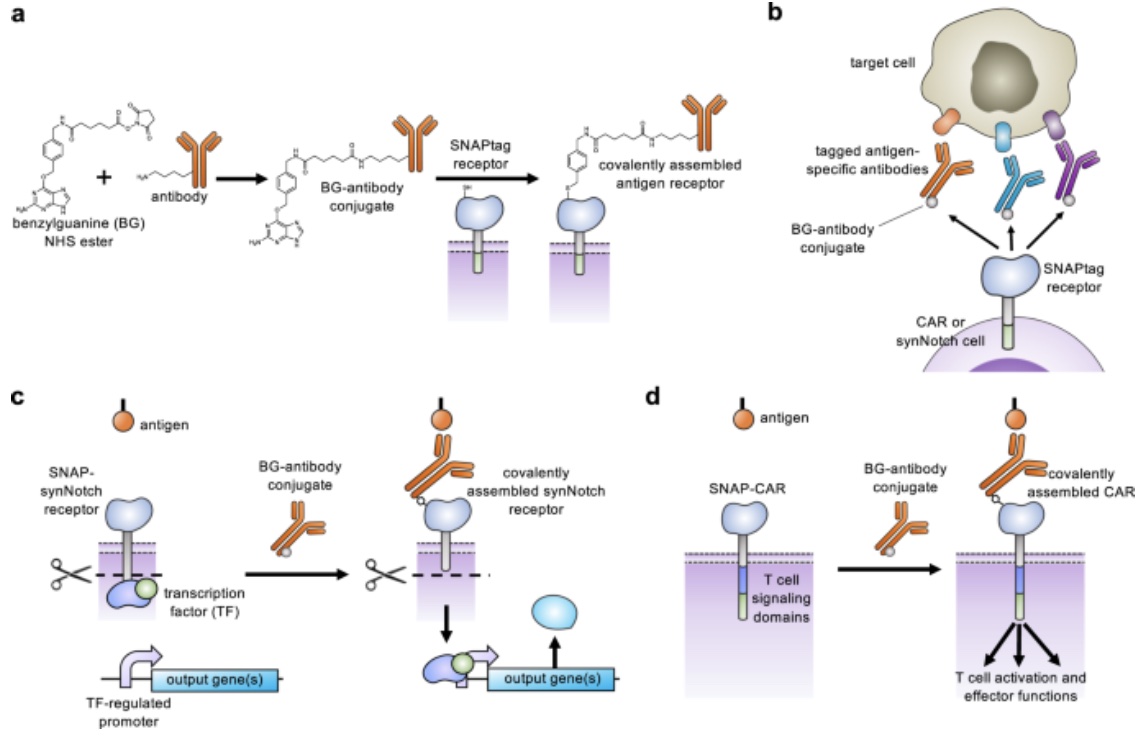
Through expanding the targeting capabilities of chimeric antigen receptors (CARs) and synthetic Notch (synNotch) receptors, University of Pittsburgh researchers have developed a “universal” receptor system that allows T cells to recognize any cell surface target. Given the potential previously seen in engineered antigen receptors, and CARs being the most clinically advanced of these technologies, the team of researchers began investigating a method to gain additional control over CAR function. By engineering T cells with receptors bearing a universal “SNAPtag”, CARs bind to the common tag molecule fused to the antigen-specific antibody, instead of directly binding antigen targets. This new approach allows for great expansion to antigen targeting and shows great promise in using CARs against additional immune-related diseases and other types of cancer.
New Research Shows That Bacteria Get “Hangry,” Too
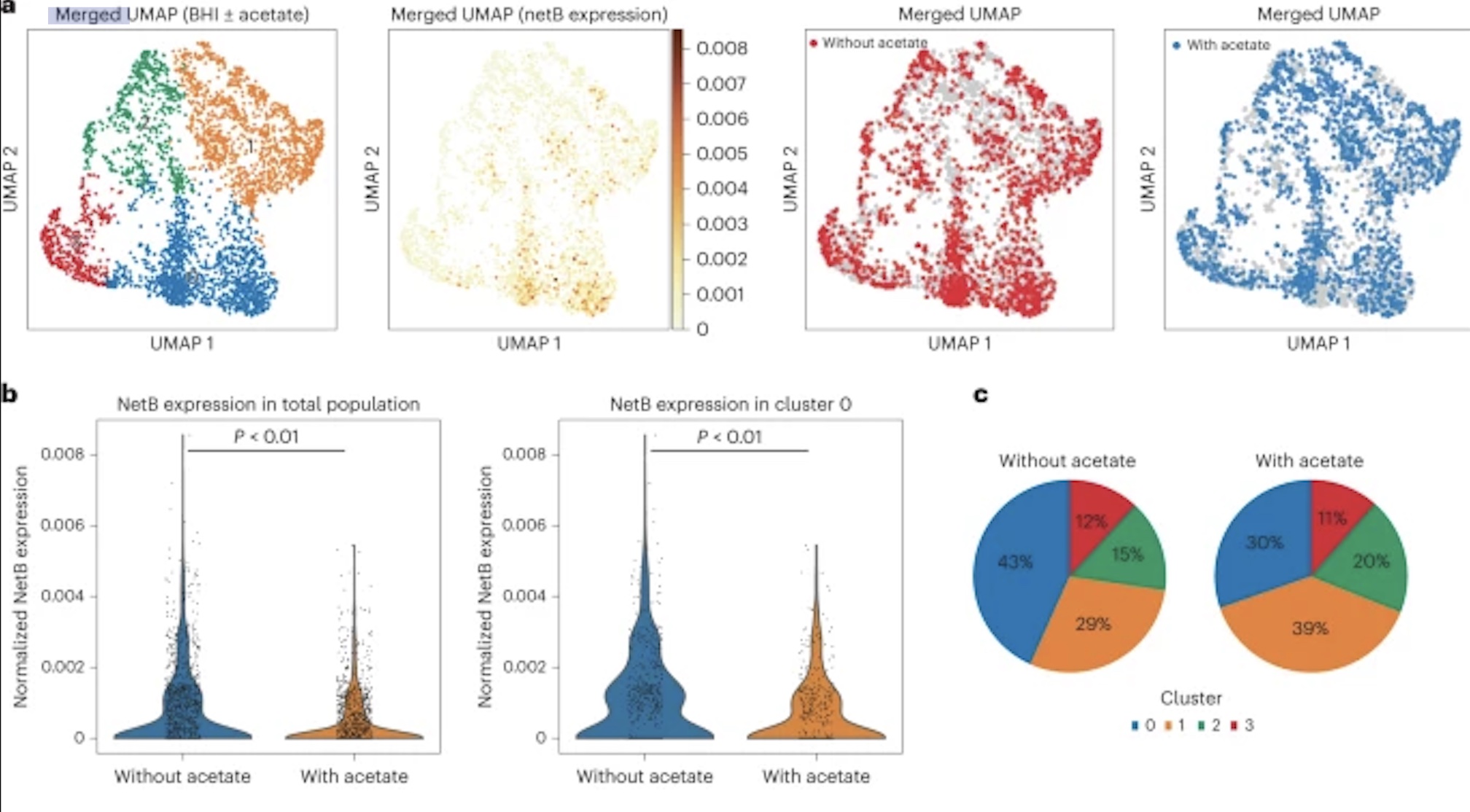
New research by Adam Rosenthal, PhD, assistant professor in the Department of Microbiology and Immunology at UNC, analyzed the cell states of isogenic bacterial populations via a developed probe-based bacterial sequencing (ProBac-seq) method. After confirming the sequencing method accurately identified known cell states and transcriptional heterogeneity of bacterial cells, Rosenthal et al. applied ProBac-seq to Clostridium perfringens and found that despite transcriptional heterogeneity, these identical cells behaved differently with varied functions within the bacterial population. Specifically, through single-cell analysis on C.perfringens and graph-based clustering, four distinct subpopulations were revealed, with all clusters preferentially expressed netB, a toxin responsible for necrotic enteritis disease. To investigate mechanisms affecting expression rates of the toxin, C.perfringen was treated with sodium acetate. Following treatment, the expression of netB was significantly less in comparison to the control cluster. These findings are just the beginning to understanding bacterial populations and mechanisms behind single-cell behavior and could lead to a new approach to antibiotic tolerance down the line.
A swapped genetic code prevents viral infections and gene transfer
In a recent study published in Nature, researchers from Harvard Medical School report on a new E. coli strain resistant to viral infections but unable to release its modified genes, effectively reducing the risk of incorporating the modified genetic material into natural cells. How? Through swapping the genetic code of an E.coli strain via viral transfer RNAs (tRNAs). The team started with the synthetic bacterium Syn61?3, which has a reduced number of codons, previously generated in 2021 by UK researchers. After preliminary experimentation, it was found the original strain was susceptible to some bacteriophages and researchers decided to implement viral tRNAs as a mechanism to establish an artificial genetic code, hoping to make the Syn61?3 strain resistant to these bacteriophages. Through efficient codon reassignment and swapping of amino acids via viral tRNAs, the E. coli strain is resistant to viral infections as viral proteomes are mistranslated and is unable to release synthetic genetic information. This study provides a new concept for future applications beyond E. coli and greater commercial potential for synthetic organisms.
James Adeosun is a student at Clare College of the University of Cambridge who spent the summer working in the lab at VIC.
This summer, I was fortunate enough to spend 11 weeks at the Vaccine and Immunotherapy Center (VIC) in Boston, immersing myself in immunology research. The VIC is directed by Professor Mark Poznansky, whose enthusiasm for science emanates around the lab. The placement was absolutely fantastic! From a research standpoint, the work seemed to be right on the cutting edge of immunology. The project I was working on centered around their self-assembling vaccine technology, mainly in the context of a therapeutic HPV vaccine. Specifically, I was testing whether using microneedle arrays was a viable method of vaccine administration, whilst also developing an in vitro assay for testing both this and other vaccines. We also theorized a new vaccine model based on some literature from a Chlamydia trachomatis vaccine trial at Hammersmith. Additionally, I got to go to the main MGH campus and see a few patients with Prof. Poznansky, which was very exciting as a medical student; the hospital was incredibly impressive. Beyond the science, the lab environment was very inspiring, with people from many different backgrounds working towards similar aims. Frequent lab events meant that I felt very welcomed from the start, and very involved by the end. For example, I was asked to arrange weekly seminars for the intern cohort, led by principal investigators of the various labs within the VIC. I was also afforded an in-depth insight into some of the technicalities of research, such as the grant writing process, as well as the possibility of having a mixed clinician / research role. The city of Boston itself had lots to offer in the downtime. Excellent public facilities and social venues provided a great cultural experience. Highlights included the famous Boston Freedom Trail, as well as eateries serving variations of the popular local hit, clam chowder. Furthermore, Boston is closely apposed to many important tourist destinations. This allowed me to travel to both Cape Cod and NYC on separate weekend trips, providing a more rounded cultural experience of the USA. In summary, the placement afforded me the opportunity of a lifetime, both academically and in terms of broader life experience, and for that I am incredibly grateful.


Celebrating the 20th Year of Annual Research Program at VIC for Clare College Students
Prof. Mark Poznansky is the Director of the Vaccine & Immunotherapy Center (VIC) at Massachusetts General Hospital and Harvard Medical School in Boston, which he founded to accelerate the discovery and development of new medical products for specific types of cancers, infectious and immune mediated diseases. One key part of VIC’s mission is the training of the next generation of innovators and scientists. Each summer Prof. Poznansky, who gained his PhD while at Clare College, provides stipendiary support for Natural Sciences or Medical students from Clare to spend 12 weeks at VIC under the supervision of a senior scientist. This annual program was initiated with the eminent Dr. Celia Duff at Clare and has been continued in a wonderful collaboration with Prof. Jason Carroll. While here at MGH, the Clare student receives extensive training in cutting edge experimental techniques and analysis of data and frequently gains coauthorship on publications ensuing from the original research conducted in the lab. Often times the selected Clare student has continued to be involved with the research remotely long after the completion of their studentship. Some Clare students requested to return for a second “tour of duty” at VIC the following summer. All the selected students from Clare College have been exceptional and many have gone on to successful careers in medicine and scientific research and report that the studentship changed their lives in pursuing a career in academia. We are now celebrating the 20th year of this very successful program that included former and current Clare students – from James Smith and Emma Anderton in 2002 and 2003 all the way to James Adeosun in 2022.
FASEB Journal publication from VIC’s Dr. Susan Raju Paul
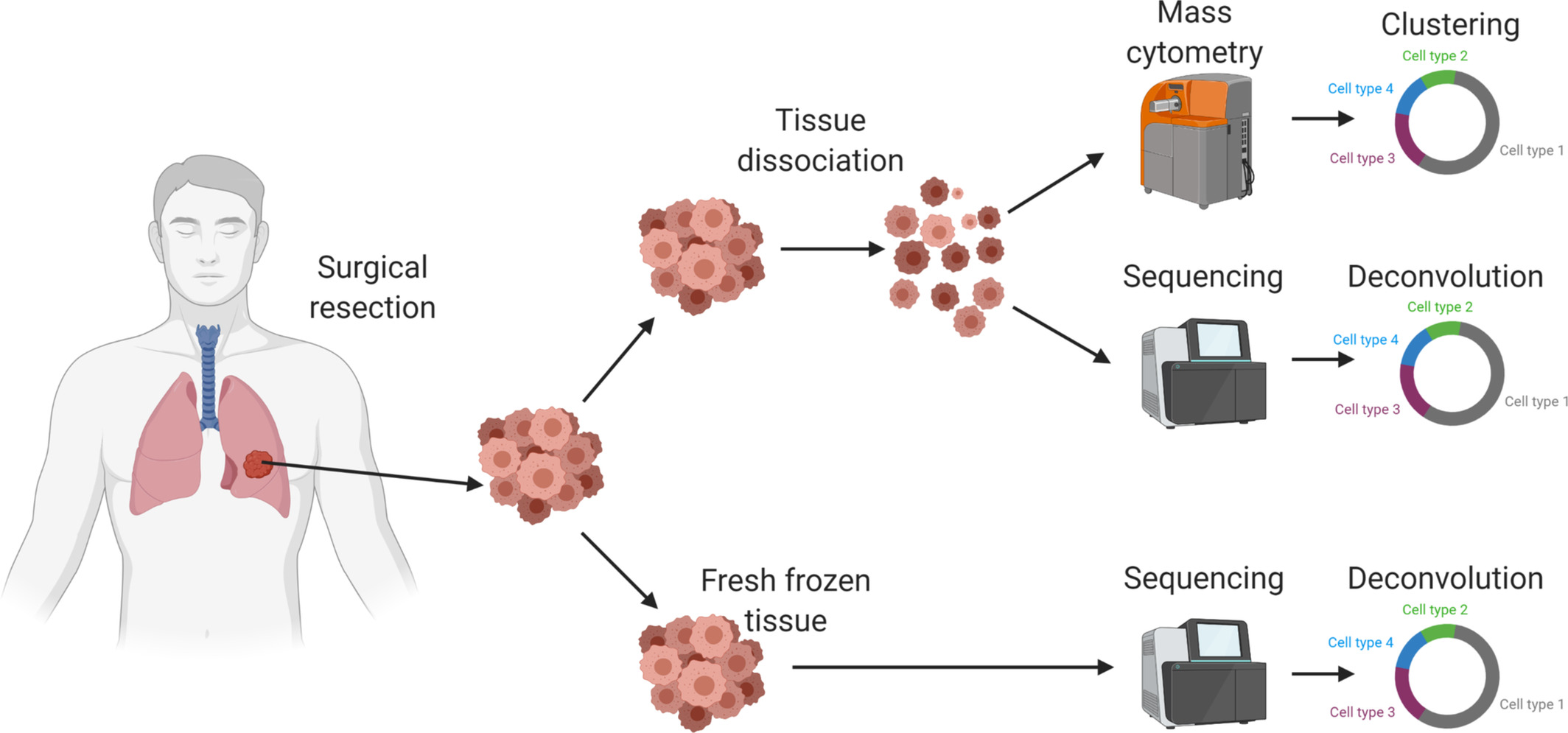
Susan Raju Paul MBBS, researcher at VIC, recently published results of an in-depth analysis of the tumor microenvironment (TME) using a small cohort of patients with non-small cell lung cancer (NSCLC), the most common histopathological variant of lung cancer. Results of the pilot study support the use of a patient’s unique TME and immune subpopulations as prognostic indicators for selection of appropriate treatment for improving patient health care outcomes. Future research lies in validating these findings with a larger cohort in effort to better characterize and understand expression of immune markers and subtypes of NSCLC and implications for treatment.
NY Times Opinion Piece from Dr. Michael Callahan:
Dr. Michael Callahan is a Staff Physician at MGH in internal medicine and infectious disease and the Director of clinical translational research at VIC. Since 2002, Dr. Callahan has investigated disease outbreaks in Asia. He recently published a piece in the NY Times covering the implications of China’s end to their zero COVID policy and how the U.S. could indirectly help the country avoid COVID catastrophe.
Bifunctional cancer cell–based vaccine concomitantly drives direct tumor killing and antitumor immunity
Despite the extensive research conducted on cancer vaccines and related therapeutics, glioblastoma remains uncured. Treatment options fall into a combination of surgery, radiation, and chemotherapy in an effort to slow tumor growth. In a promising new study led by researchers from Brigham and Women’s Hospital, potential is seen in a new cell therapy that contributes to both the elimination of current tumors and prevention of tumor recurrence, seen in mouse models of glioblastoma. The new cell therapy repurposes live tumor cells with gene engineering, altering these cells to release dual cell-killing and immunomodulatory agents, capable of killing tumor cells and stimulating the immune system for further cancer prevention.
Optimized ACE2 decoys neutralize antibody-resistant SARS-CoV-2 variants through functional receptor mimicry and treat infection in vivo
As a result of evolution under the selective pressure of early-stage pandemic therapeutics and vaccines, plus natural immunity, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is becoming endemic. Concerns rise as SARS-CoV-2 variants continue to emerge with resistance to currently available monoclonal antibody therapeutics. Recent research led at Dana-Farber Cancer Institute and with direct involvement of the VIC team at Mass General Hospital, investigates the potential in developing a decoy drug therapy that may be capable of treating common and arising variants of SARS-COV-2. The decoy drug uses angiotensin-converting enzyme 2 (ACE2) decoy receptors, previously proposed as a therapeutic for COVID-19. These ACE2 decoy receptors engage the receptor binding domain of the viral S protein and outcompete human cell-surface viral binding, effectively neutralizing viral infection. Also, as the viral binding domain of the ACE2 decoy is identical to cell-surface ACE2, resistant variants are not likely to emerge as any mutations that reduce affinity of the virus for the decoy reduces viral infectivity. Research findings provide new insight to ACE2 decoy functionality and indicate potential in further development as a therapeutic for ACE2-dependent coronaviruses.